Introduction and Research Perspective
The goal of our research program is to discover how system-level properties of large groups of eukaryotic and prokaryotic cells emerge from microscopic dynamics, both at single-cell and molecular scales. Single cell behavior is well understood in eukaryotes and prokaryotes, but it is their behavior in large communities that is important to multi-cellular life. For all eukaryotic and prokaryotic multi-cellular organisms, life depends on the coherent functioning of large cell aggregates. In traditional condensed matter physics, great progress is made by discovering how bulk system properties arise from the behavior of the sub-units. If the same is true of living condensed-matter, then an understanding of the relationship between the cellular sub-units and the function of their collections is crucial to understanding life in multi-cellular organisms and, as a consequence, controlling it.
Large cell groups that function collectively, such as tissues and bacterial biofilms, often have sub-groups with specialized jobs. This heterogeneity in cell function is most dramatically illustrated by comparing cells at the surfaces to cells within the bulk. For example, in stratified epithelia, the apical and basal layers of cells have totally different functions than the inner lateral cells. Likewise, in bacterial biofilms, cells at the interface between the colony and nutrients are always the most metabolically active, and cells far away from nutrient sources must rely on the metabolic intermediates of surface cell groups. Thus, to understand collective behavior of multi-cellular systems as wholes, our research program focuses on eukaryotic and bacterial cell layers on surfaces, as well as many types of bulk three-dimensional cell aggregates.
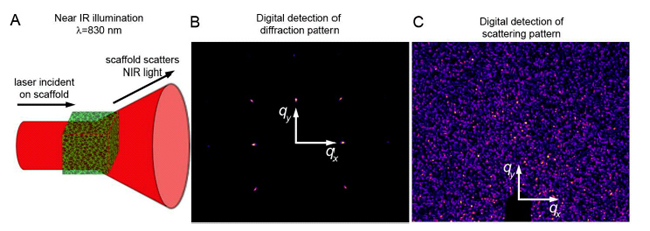
It is impossible to see deep into thick 3D cell aggregates with visible light due to strong multiple scattering and absorption. To surmount these imaging problems, we are building new tools that use radiation at wavelengths that pass through biological materials with, primarily, single-scattering events: x-rays and IR illumination. These tools are currently being developed and, thus, are top-secret. If you would like to see them in action, come pay us a visit!
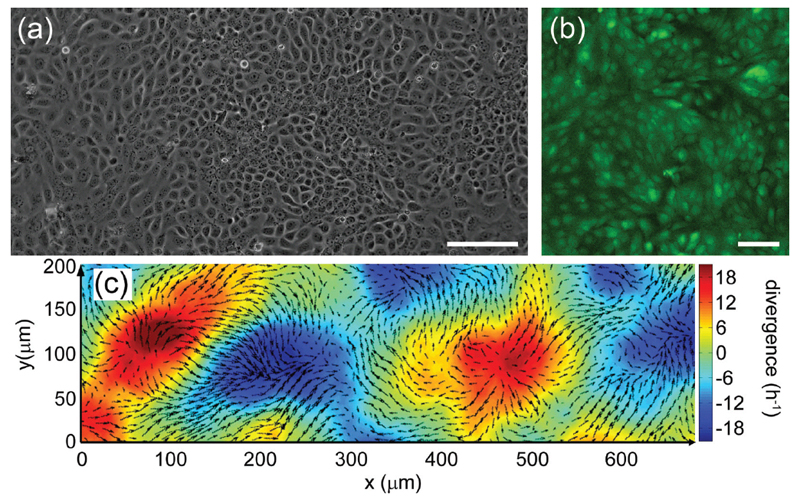
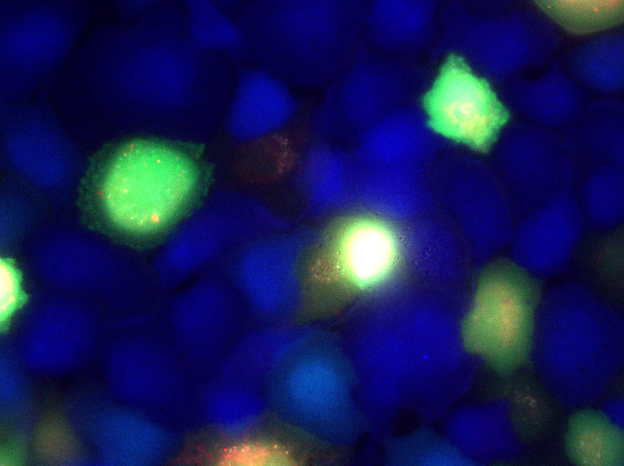
We also study cells on surfaces, which can be imaged with visible light, allowing the use of ordinary fluorescence microscopy. Throughout our research program, we compliment micro-scale studies macro-scale measurements involving low-resolution time-lapse imaging and bulk-rheological studies. At the molecular-scale, we do x-ray scattering to explore bio-molecular self-assembly to understand the interactions between the many extracellular materials, as well as their interaction with cell surfaces.