My research

Population genetics of the central Florida scrub

For my dissertation I did a comparative phylogeographic analysis of both plant and animal taxa endemic to the ridge comparing origin (based on the range of the closest relative) and age of divergence. Common patterns can shed light on the origins of the high endemism on the ridge as well as mechanisms of speciation. I tested between two phylogeographical hypotheses for each of my five species’ data sets. The first hypothesis supports a more ancient origin of the central Florida scrub ecosystem, with migrations of species coming from the West, and the second hypothesis supports a more recent origin of the system, when central Florida was a refuge for many present-day northern species during the last glaciations. All fieldwork, phylogenetic lab work, and analyses have been completed, and I am currently working on the preparation of the manuscript for these results (expected to be submitted to the Journal of Biogeography in March/April). DNA sequences for the ITS region of the nuclear ribosomal RNA genes were generated for several accessions of each Florida endemic species and its putative sister species using universal primers, or more taxon-specific primers in the case of Persea. For Persea and Prunus, several regions of the plastid genome were added in an effort to gain resolution.
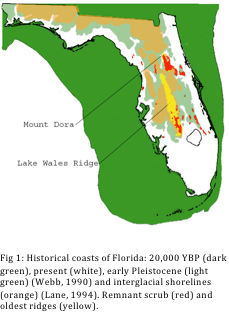
Low diversity can lead to a reduced ability to respond to current and future environmental changes and evolutionary pressures, increased risks of inbreeding and drift, and ultimately extinction. In general, rare species are believed to have a lower genetic diversity than widespread species, and this may be especially true for a species that has recently experienced habitat loss and population declines. However, the causes (Baskauf, Eickmeier, 1994) and forms (Rabinowitz, 1981) of rarity, species age (Fernald, 1926; Willis, 1922) and phylogenetic history (Gitzendanner, Soltis, 2000) have all also been shown or hypothesized to affect genetic diversity in rare species. To maximize our understanding of genetic diversity in the rare species of the central Florida scrub, my dissertation work compares the diversity of five endemic species to their more common sister species.
Genetic variation within one or several species in a system is increasingly recognized as a direct indicator of ecosystem health, ability to face present and future evolutionary pressures, and the diversity of other species in the system (Mitchell-Olds et al., 2007; Vellend, Geber, 2005; Wade, 2007; Whitham et al., 2003). Any species-specific or common pattern of genetic diversity we find will therefore be useful for our understanding of the conservation needs of the central Florida scrub. Different approaches to measuring genetic diversity will be explored, such as the differences between classic heterozygosity, percentage of polymorphic loci, and the more recent idea that allelic richness is a better identifier of conservation priority areas, emphasizing unique alleles in a population. The study of gene flow will help in identifying pathways that could form the basis of corridor design, or combining protected areas, to enable mixing of populations that were previously sympatric but are now fragmented. A refined conservation management plan will be recommended to different organizations through presentations and publications. This study is providing the first genetic information for these species, and will help us understand their dynamics, differences and commonalities.




Polyploidy in Asimina obovata
After finding more than two alleles per locus for several Asimina obovata individuals during the screening of the microsatellites, we investigated the ploidy of a sample of those plants. In order to do so, we went back to some populations and, after obtaining authorization, took whole plants to grow in the greenhouse. Root tip squashes reveal the presence of dipoid individuals (2n=18) in populations where only 2 alleles were observed per locus, as well as in nursery plants. In populations where 3 or 4 alleles were observed in more than one locus, root tip squashes from several plants from one of the populations reveal the presence of both triploids (2n= 27) and tetraploids (2n =36). Also, flow cytometry methods have been optimized in order to estimate ploidy (102). With this method, dried leaf tissue can be used and all samples are being screened in order to determine their ploidy leel.
The central Florida scrub is a unique environment with a unique history, and the different ploidal levels in the endemic Asimina obovata has important potential implications both for the conservation of the species, and for the advancement of our understanding of the role of polyploidy. Another species endemic to the Lake Wales Ridge, Crotalaria avonensis, has also recently been discovered to be a polyploid (Gitzendanner, unpublished). The implication of such discoveries can be crucial for this ecosystem and should be investigated more fully.
The implications of polyploidy are important as a source of evolutionary and ecological novelty, potentially resulting in habitat expansion outside of the progenitor’s population limits, or adaptation to future changes. Polyploidy has seldom been discussed in a conservation context, but some authors argue for special consideration of polyploid populations in their own right, or for separate conservation plans for populations with each level of ploidy.


Other work on the central Florida scrub
I worked with Dr. Matt Gitzendanner on Ziziphus celata, the rarest plant in Florida. My former undergraduate student Patricia Soria and I developed a microsatellite library for this species. We found some genetic diversity where other markers had failed. This is especially important for this species as all common garden and manual pollinations have failed for this plant to produce any viable seed. Without any reproduction, this species is domed. Matt Gitzendanner is also working on the self-incompatibility allele markers in order to facilitate future population augmentation in the field.
A paper is iwas submitted to Conservation Genetics on the microsatellite data results
Sustainable logging of Mahogany
Tropical forests are one of the most threatened ecosystems, and logging is one of the main threats. true mahogany (Swietenia macrophylla King.) has been the target of abusive exploitation for centuries and is the most commercially important tropical timber on the international market. The species is commercially extinct within most of its range and protected in some areas; such as the study area Wet Botes, Rio Bravo, in Belize. Sustainable logging is being tried in the hope of finding a compromise between conservation and market demand. West Botes is a plot that has been selectively logged and the genetic diversity needs to be assessed to evaluate the actual sustainability of the logging regime.
The aim of this project was to use molecular markers to assess the spatial genetic structure of the population of Swietenia macrophylla, its genetic diversity, mating system and gene flow. To do so, three different microsatellites were used on both samples from the adult trees and also on the seeds in the plot.
We found that the spatial genetic structure of the population revealed tight clustering of closely related trees and a significant level of biparental inbreeding. However, we found out once the experiment had started that the West Botes was recovering from past intense logging and one hurricane, so that the negative impact might not have come from the logging regime. The low number of loci might also have biased the results.



In my current post-doctoral position, I am using phylogenies to address questions of large-scale plant community assembly and conservation.
I am helping to assemble a tree encompassing all plant species of Florida (~4,100 taxa), along with georeferenced locations for most of them (~168,000 points for > 3,000 of the species). I am using GIS layers to build a species distribution model (using MaxEnt) that will enable us to infer the biotic and abiotic factors influencing the current distribution of entire communities in Florida and predict their distribution based on different future climate models.
Also, the historical record from herbarium specimens will enable us to identify recent shifts in distribution of species. This analysis will help to better predict the changes of plant distributions in the face of increasing urbanization and climate change and which taxa may me most susceptible to environmental changes.
The Florida phylogeny
